There was a bit of controversy that played out earlier this year on arxiv and the pages of MNRAS on a subject of interest to me, namely full spectrum modeling of (simulated) galaxy SEDs. It began with a paper by Ge et al. (arxiv article ID 1805.03972) comparing outputs of STARLIGHT and pPXF, two popular full spectrum fitting codes, on very simple one and two component synthetic spectra with varying ages, reddening, and S/N. They found significant biases in mass to light ratios and other derived quantities in STARLIGHT output in some cases — primarily ones with a young population and large reddening — that oddly got worse with increasing S/N, while pPXF generally had smaller biases that improved with increasing S/N. They also obtained poor fits to the data in the cases with the most biased outputs, and noted multiple order of magnitude differences in execution time.
Not surprisingly this was soon followed with a heated rebuttal by cid Fernandes, the primary author and maintainer of STARLIGHT. This was first posted on arxiv with the title “Rebutting fake news on full spectral fitting” (article ID 1807.10423). The published version with the more temperate title “On tests of full spectral fitting algorithms” appeared in the November 2018 MNRAS (and is apparently not paywalled, although Ge et al. is).
As I read it there were three main threads to the rebuttal:
- A single, “hitherto deemed unimportant” line of code was responsible for the poor performance at large input reddening values. The offending line apparently limits the initialization of reddening values to what would normally be reasonable for realistic galaxy spectra.
- But, even without modifying the code good results could have been achieved by modifying configuration files.
- And anyway the worst performance was for an unrealistic edge case that wouldn’t be encountered in real work.
The one problem I have with this is that STARLIGHT is not open source at present, and without the source code the otherwise trivial issue isn’t easily discoverable, let alone fixable. As for the other points it should have been fairly obvious that getting poor results when the ingredients used to fit a data set are exactly the ones used to generate the data is a sure sign of convergence failure.
Actually though this post isn’t about these papers or directly about these two codes. It wasn’t clear to me why Ge et al. wrote the paper they did or why editors and referees considered it publishable in MNRAS — while fake data exercises are a necessary part of software validation there wasn’t any larger scientific purpose to their work. But I did like the idea of looking at an unrealistic edge case for reasons that I hope will become obvious. What I’m going to examine is even simpler: I construct a mock spectrum as a linear combination of two SSP model spectra from the EMILES library, perturb it with random noise, and fit it with a subset of EMILES using three different fitting methods. Neither convolution with a line of sight velocity distribution nor reddening is done, and these aren’t modeled in the fits. The fitting methods I use are
- non-negative least squares using the R package
nnls, which implements Lawson & Hanson’s 1974 algorithm. This is a crucial part of both my code for maximum likelihood estimation of star formation histories andpPXF, which uses the implementation (probably based on the sameFORTRANcode) in scipy.optimize. - A Stan model that’s a simplified version of the one I use for Bayesian SFH modeling.
STARLIGHTuses a Monte Carlo based optimization scheme, and I thought it would be interesting to compare the methods I use regularly to a state of the art approach to Monte Carlo optimization. To that end I chose the “differential evolution” algorithm implemented in the package RcppDEoptim. I should emphasize at the outset that this is a very different algorithm than the one inSTARLIGHTand none of the results I present here have any direct relevance to it.
I didn’t think it was worth the effort to version control this or create a Github project, but I did upload all code and data needed to reproduce the graphics in this post to my dropbox account. I don’t necessarily recommend running this because it’s quite time consuming, but if you want to try it just download the entire folder and after starting R set the working directory to toynn and type source("toynnscript.r", echo=TRUE). Then go away and come back in 12 hours or so.
Let’s look at the script. The first few lines load the libraries I need and set some options for rstan:
## libraries we'll need
require(rstan)
require(nnls)
require(RcppDE)
require(ggplot2)
require(bayesplot)
require(gridExtra)
source("toynnutils.r")
## set a few options
ncores <- min(parallel::detectCores(), 4)
options(mc.cores = ncores)
rstan_options(auto_write = TRUE)
set.seed(654321L)Stan can run multiple chains in parallel on any multi-core architecture and defaults to 4 chains, so the line that starts with ncores <- will be used to tell Stan to use the smaller of the number of cores detected or 4. I set and reset the random number seed several times in this script for exact reproducibility. Next I set up the fake data:
## load emiles ssp library and extract parts we need
load("emiles.sub1.rda")
X <- as.matrix(emiles.sub1[, 110:163])
X <- scale(X, center=FALSE)
mstar <- mstar.emiles[109:162]
ages <- ages.emiles
lambda <- emiles.sub1[,1]
rm(emiles.sub1, mstar.emiles, ages.emiles, Z.emiles, gri.emiles)
nr <- nrow(X)
nv <- ncol(X)
beta_act <- numeric(nv)
beta_act[22] <- 0.3
beta_act[50] <- 1 - beta_act[22]
sdev <- 0.02
y_act <- as.vector(X %*% beta_act)
y <- y_act + rnorm(nr, sd=sdev)
dT <- diff(c(0, 10^(ages-9)))
stan_dat <- list(N=nr, M=nv, X=X, y=y, dT=dT, sdev=sdev)The subset of the EMILES library provided here has 4 metallicity bins and 6000 wavelength points between about 3465 and 8864Å. I extract the solar metallicity model spectra, and although it’s overkill for this exercise I retain all 6000 wavelength points. The two nonzero components are 1 and 12Gyr old SSPs. I add IID gaussian random noise to the model spectrum with a standard deviation that will be treated as known. Real modern spectroscopic data always comes with an error estimate for each flux measurement, and these are typically treated as accurate in SED modeling.
The script runs the Stan model next, so let’s look at the Stan code:
data {
int<lower=1> N;
int<lower=1> M;
matrix[N, M] X;
vector[N] y;
vector<lower=0.>[M] dT;
real<lower=0.> sdev;
}
parameters {
vector<lower=0.>[M] beta;
}
model {
beta ~ normal(0, 4.*dT);
y ~ normal(X * beta, sdev);
}
generated quantities {
vector[N] log_lik;
vector[N] yhat;
vector[N] y_new;
vector[N] res;
real ll;
yhat = X*beta;
for (n in 1:N) {
log_lik[n] = normal_lpdf(y[n] | yhat[n], sdev);
y_new[n] = normal_rng(yhat[n], sdev);
res[n] = (y[n] - yhat[n])/sdev;
}
ll = sum(log_lik);
}There are just two lines in the model block: one to specify a prior and one for the likelihood. The prior was copied directly from my “real” Stan model for SFH modeling. It sets the scale parameter to be proportional to the width of the age bin each SSP model covers, and since the parameter values are proportional to the mass born in each age bin this will make a typical draw from the prior have a roughly constant star formation rate over cosmic history. Unfortunately I rescale the data in a different way for this exercise and the prior is doing something different, but no matter for now. Continuing with the script:
## in the stan call set open_progress to FALSE if using rstudio
## max_treedepth = 13 should make convergence warnings go away
## this will take a while...
system.time(stanfit <- stan(file="simplenn.stan", data=stan_dat, chains=4, iter=1000, warmup=500,
seed=54321L, cores=ncores,
open_progress=TRUE, control=list(max_treedepth=13)))I set the number of warmup and total iterations to half the rstan defaults, which is plenty. Stan gets to stationary distributions for all parameters remarkably quickly. In the comments I optimistically guess that all convergence warnings will go away. Actually there was a warning that “17 transitions exceeded (sic) the maximum treedepth.” In this case the warnings don’t actually point to any convergence issues. BTW this took about 1 1/4 hours on a 2018 vintage PC built around Intel’s second most powerful consumer CPU as of late 2017.
In the paper that introduced pPXF Cappellari and Ensellem (2004) had the important insight that non-negative least squares produces the maximum likelihood estimator for the stellar contributions in SED fitting (in astronomers’ parlance it minimizes χ2). Solving NNLS problems is important in a number of disciplines and many algorithms exist, several of which are implemented in various R packages. It turns out though that an old one published by Lawson and Hanson works pretty well for the type of data used in SED modeling. A remarkable feature of NNLS is that it tends to return sparse (or parsimonious as I put it in an earlier post) parameter estimates. In fact with real data I find that it always returns sparse solutions for the stellar components. Getting back to the script the last major computational efforts involve repeated calls to nnls and a smaller number of simulations using DEoptim (which is much slower):
## simulate lots of nnls fits
system.time(nnfits <- sim_nn(X, y_act, sdev=sdev, N=2000))
## nnls likes to produce sparse solutions, but not so sparse as the input in this case
qplot(nnfits$simdat$nnzero,geom='histogram',binwidth=1) +
geom_vline(xintercept=median(nnfits$simdat$nnzero)) +
xlab("# components")+ggtitle("# components in NNLS fits")
table(nnfits$simdat$nnzero)
## simulate not so many calls to DEoptim. This also takes a while...
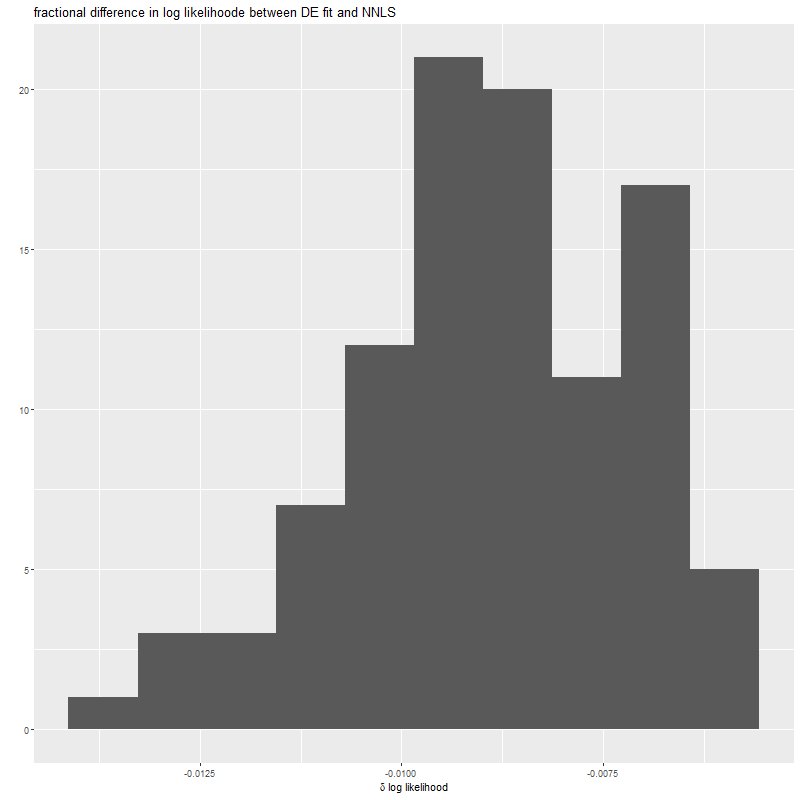
system.time(defits <- sim_de(X, y_act, sdev=sdev, N=100))
x11()
qplot(defits$simdat$deltall,geom='histogram',bins=10) +
xlab(expression(paste(delta~"log likelihood"))) +
ggtitle("fractional difference in log likelihood between DE fit and NNLS")The function sim_nn is in the file toynnutils.r. This just generates N randomly perturbed sets of the observation vector y, fits them using nnls, and returns some information about the fits (in Windows the CPU time estimates will be not very accurate):
sim_nn <- function(X, y_act, sdev=0.02, N=1000, seed=12345L) {
require(nnls)
set.seed(seed)
nnzero <- numeric(N)
ctime <- numeric(N)
ll <- numeric(N)
nr <- length(y_act)
nv <- ncol(X)
B <- matrix(0, N, nv)
res <- matrix(0, N, nr)
for (i in 1:N) {
y <- y_act + rnorm(nr, sd=sdev)
ctime[i] <- system.time(nnsol <- nnls(X,y))[1]
B[i,] <- nnsol$x
nnzero[i] <- nnsol$nsetp
ll[i] <- sum(dnorm(residuals(nnsol), sd=sdev, log=TRUE))
res[i, ] <- residuals(nnsol)/sdev
}
list(simdat=data.frame(ctime=ctime, nnzero=nnzero, ll=ll), B_nn=B, res_nn=res)
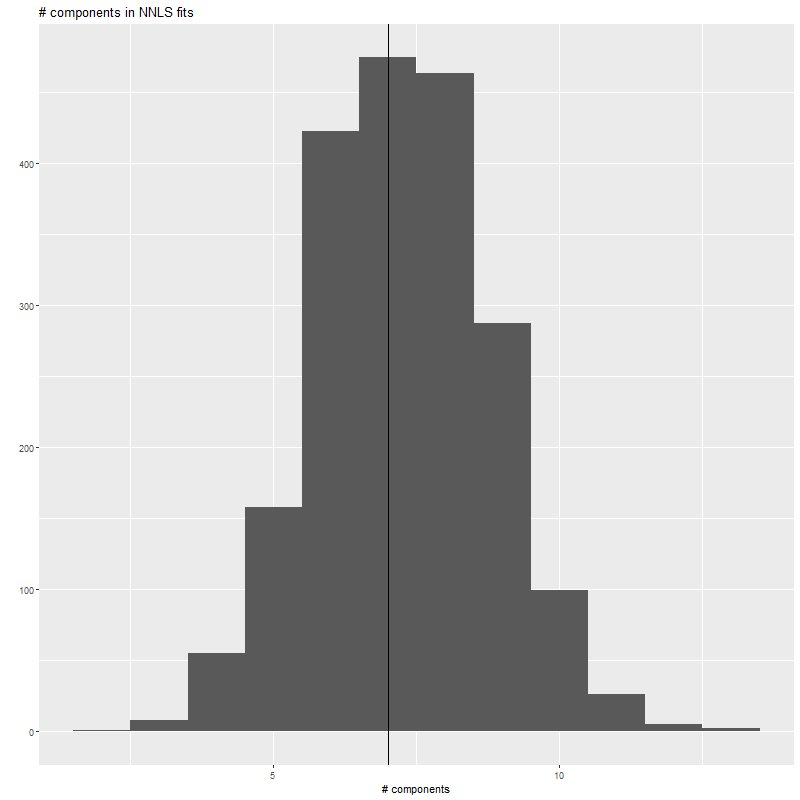
}The first ggplot2 call in the script produces this histogram of the number of non-negative components of the nnls fits. As expected and consistent with typical real data nnls always returns sparse fits, although almost never quite so sparse as the input. The call to table() returns
> table(nnfits$simdat$nnzero) 2 3 4 5 6 7 8 9 10 11 12 13 1 8 55 158 422 474 463 287 99 26 5 2
Perhaps the most interesting result of these simulations are the distributions of parameter estimates — these show 50 (thick lines) and 90% (thin lines) intervals, with median values marked by symbols:
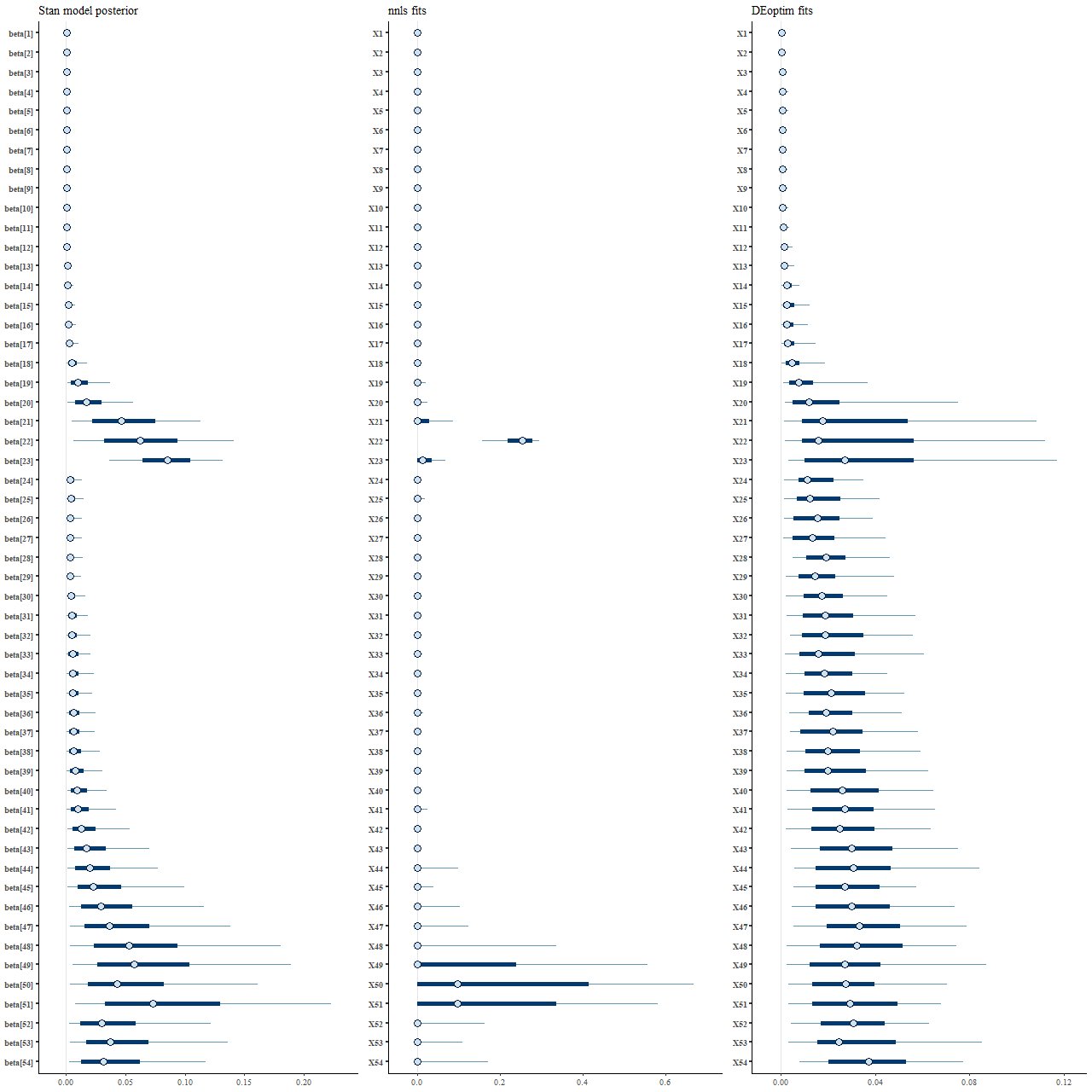
(L) posterior distribution from Stan
(M) NNLS
(R) DEoptim
What’s most striking to me at least is how different the posterior marginal distributions from Stan (left panel) are from the maximum likelihood fits (middle). Star formation is much more extended in time in the Stan model, with both “bursts” replaced with broad ramps up and decays. The Monte Carlo optimization results look superficially similar to the Stan posterior, but as I’ll show soon there are different causes for their behavior.
Let’s take a closer look at the distributions of some of the parameter values. This is called a pairs, or sometimes corner, plot — it displays the marginal distribution of each selected parameter along the diagonal and fills out the remaining rows and columns with the joint distributions of pairs of parameters. For this plot I just selected the two non-zero inputs and the surrounding age bins. The top rows are the Stan posterior, the bottom the nnls fits from the 2000 sample simulation.
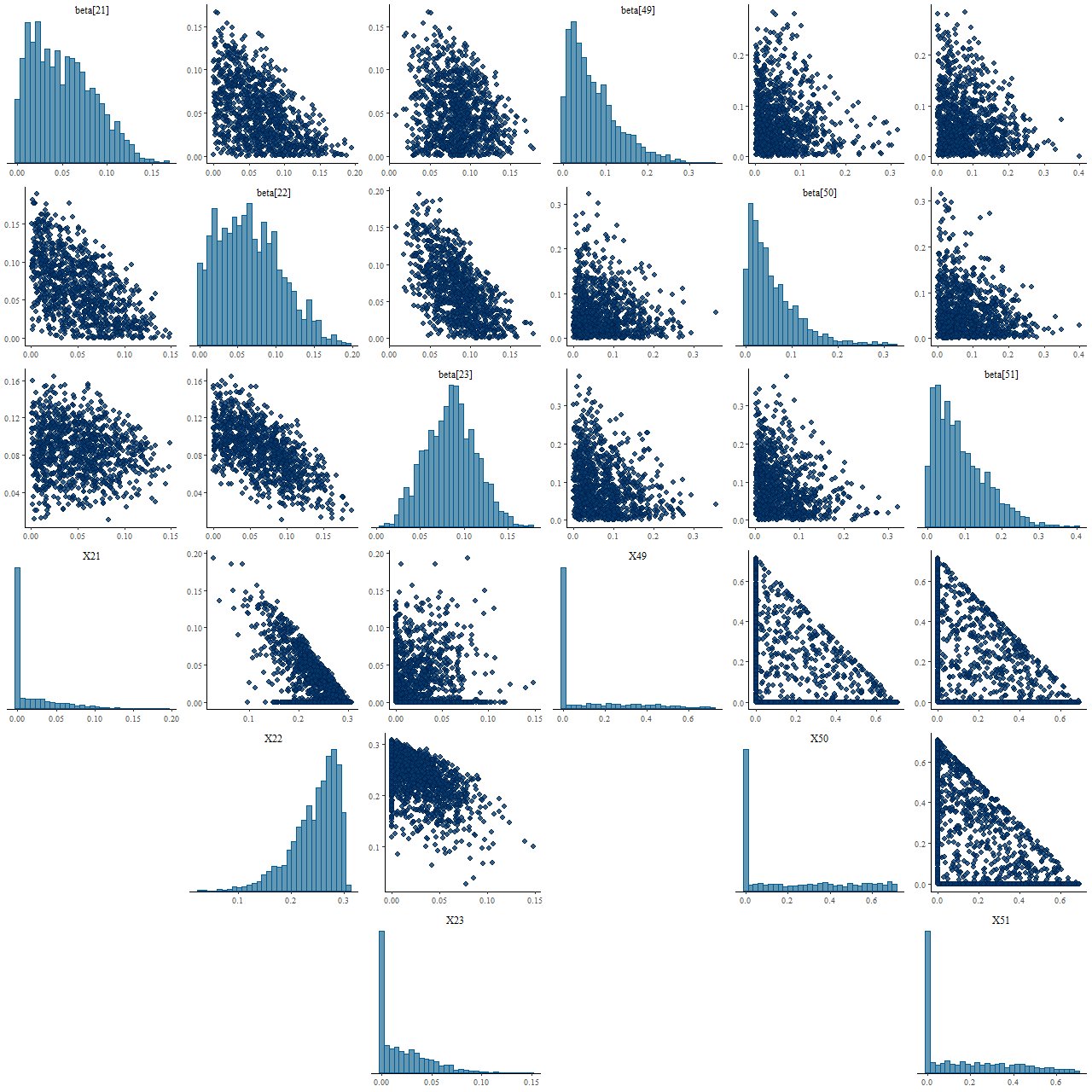
(T) Posterior draws from Stan model
(B) NNLS estimates
In frequentist statistics a confidence interval is a statement about the behavior of the estimator for a parameter under repeated experiments with the same conditions. It’s not a statement about the parameter itself, which is forever unknowable. A Bayesian confidence interval (to the fussy, credible interval) on the other hand is a statement about the parameter itself.
I’m unaware of any formula for confidence intervals of nnls coefficient estimates, but the simulation we performed gives good numerical estimates of the distribution of parameter values, and these are clearly significantly different from the posterior distributions from Stan. So, not only is there a difference in interpretation between frequentist and Bayesian confidence intervals in this case the values obtained are significantly different as well.
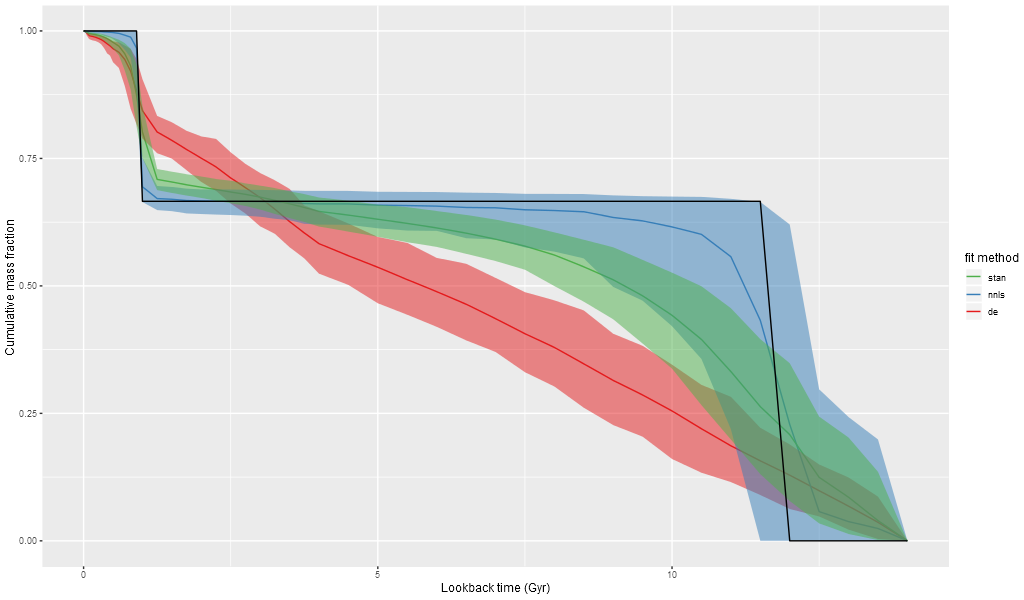
Mass growth histories have become a popular tool for visualizing SFH models, so for fun I created mock histories for the three methods tested here. These show medians and 95% confidence bands. The input MGH just falls within the 95% confidence band of the nnls fits, although the median has more extended mass growth.
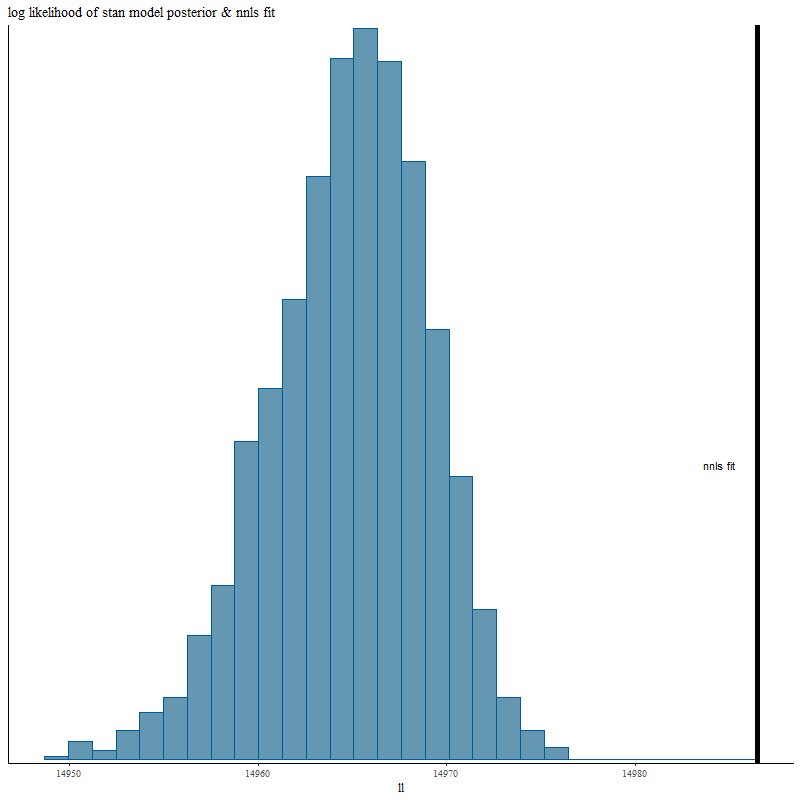
Turning next to fits to the data let’s look first at the overall fits measured by summed log-likelihood. Remember that the nnls solution is the maximum likelihood estimator and therefore neither of the other fitting methods can exceed its log-likelihood for any given realization of the data.
Here’s the posterior log-likelihood from the Stan model run. The vertical line to the right is for the nnls fit. The Stan fit average falls short of the maximum likelihood by about 0.1%. We’ll examine why in more detail next post.
I only ran 100 simulations of the “differential evolution” optimizer because it takes rather a long time (average ~450 sec. per data realization on my PC). The output log-likelihood consistently falls about 1% short of the nnls solution:
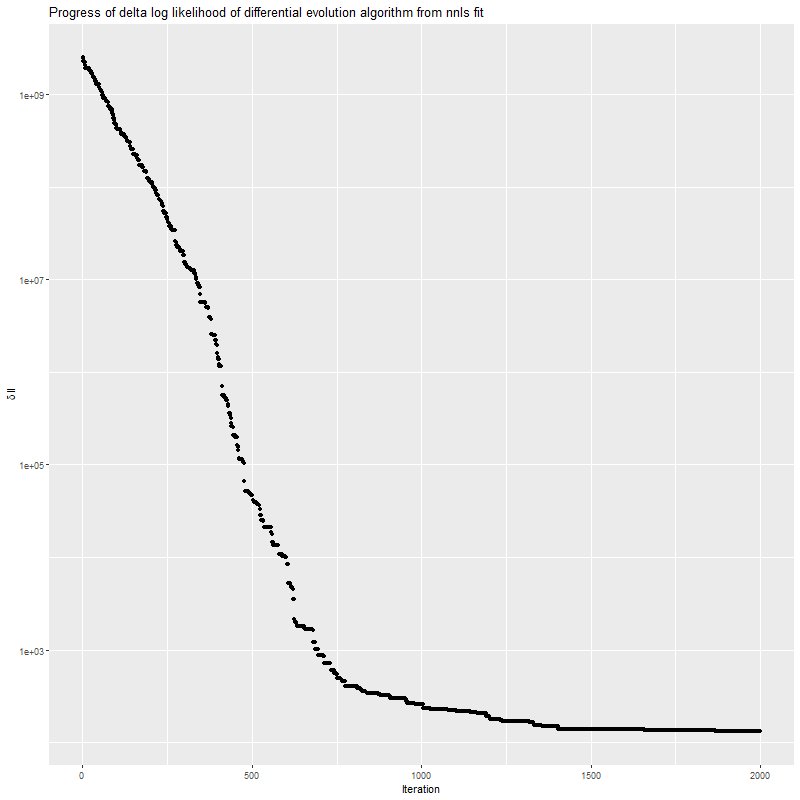
This shows the progress of the optimizer for a single data realization. Progress is rapid at first, but it slows down as it gets close to the optimum and can stall out for many iterations. This algorithm has a hard time getting to a sparse solution.
Next let’s look at detailed fits to the data. The top pane shows the unperturbed spectrum (black points) and a 95% confidence band for the posterior predictive fit (green ribbon) from the Stan model. What’s that? Simple: take parameter draws from the posterior and generate new data according to the model. This is done with the following lines in the generated quantities block of the Stan code:
yhat = X*beta;
for (n in 1:N) {
log_lik[n] = normal_lpdf(y[n] | yhat[n], sdev);
y_new[n] = normal_rng(yhat[n], sdev);
res[n] = (y[n] - yhat[n])/sdev;
}
ll = sum(log_lik);
It turns out that the 95% confidence band covers 100% of the true, unperturbed, spectral values and 95% of the perturbed ones, which indicates a successful model.
The remaining panes show several sets of normalized residuals. The first surprised me a bit at first: this shows the residuals (observed – model) of the Stan model posterior. The “points” in this plot are actually error bars — it turns out the range of residuals at any wavelength is rather small (I naively expected that residuals at each point would vary randomly around 0). The residuals from the posterior predictive fits shown in the next panel do show this behavior. This and the remaining panels display median values as points and 95% confidence bands.
The 4th pane shows the errors in the posterior predictive fits, that is the original unperturbed data minus the posterior predictive values. There are hints of systematics here: a bit of curvature at the red and blue ends of the spectrum and a few absorption lines are slightly misfit.
The final two are residuals from the simulated nnls and Monte Carlo optimization fits. The nnls fits have exactly the expected statistical behavior while the latter clearly struggles to fit the data. This is because the overly extended star formation histories reached in the number of iterations allotted result in stellar temperature distributions that aren’t quite right, affecting both the continuum and many absorption features.
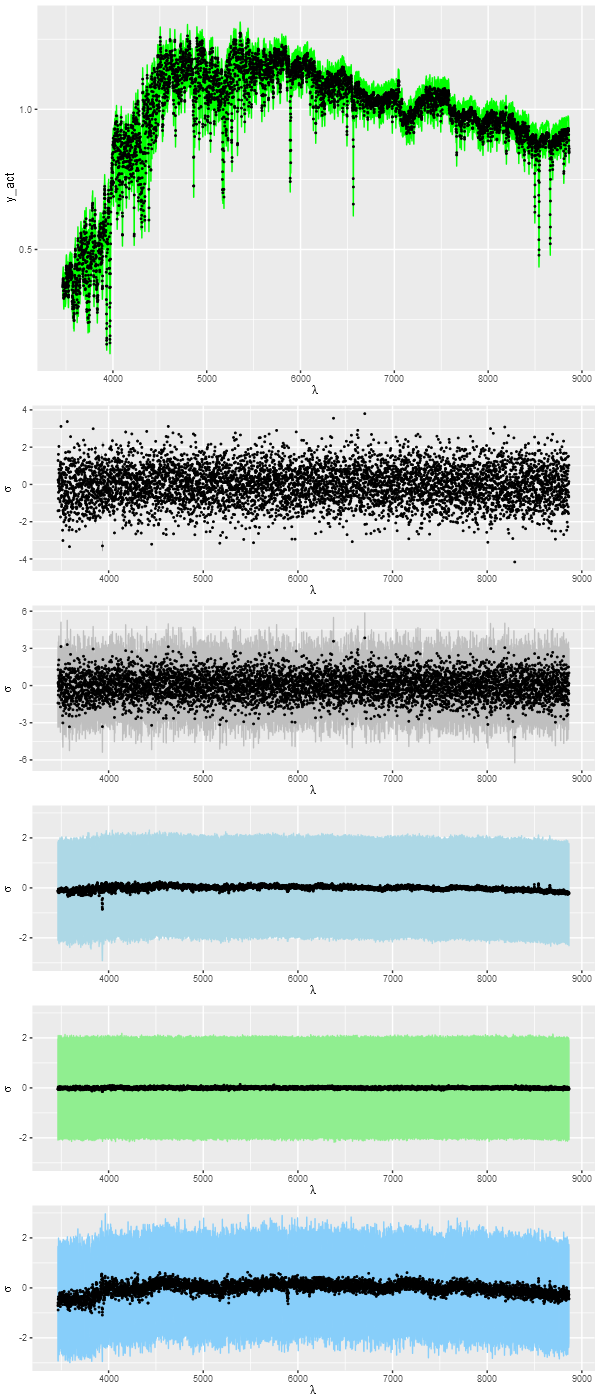
T) input spectrum and posterior predictive fit from Stan model
2) Residuals from Stan model posterior
3) Residuals from posterior predictive fits
4) Errors from posterior predictive fits
5) Residuals from NNLS fits
6) Residuals from DEoptim fits
Finally, here is a comparison of the distribution of the residuals in the Stan fit to the same data realization fit by nnls, the colored bands being the former. These show kernel smoothed density estimates on the left and empirical CDF on the right.
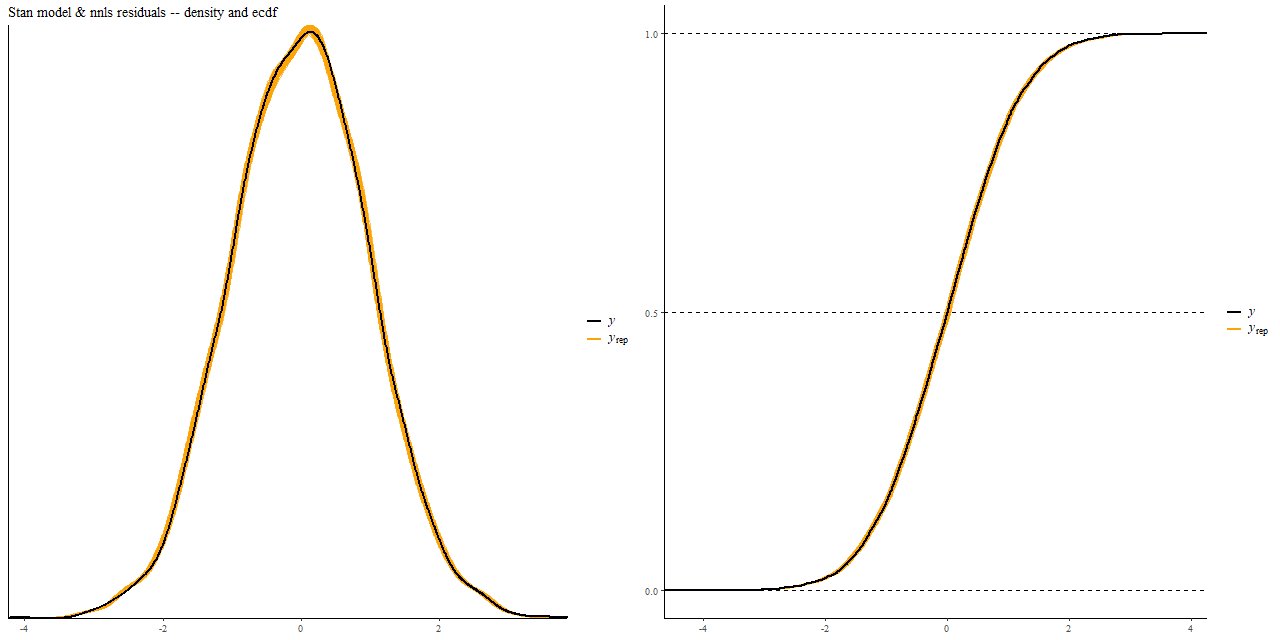
It’s sometimes suggested in the SFH modeling literature to get parameter uncertainty estimates by randomly perturbing data and refitting it. This is a computationally viable strategy with a competent non-negative least squares solver, but as we saw above this is likely to produce very different estimates than a proper Bayesian analysis.
